A foundation model for continuous glucose monitoring data
TL;DR
GluFormer is a generative foundation model for continuous glucose monitoring (CGM) data that improves prediction of glycaemic outcomes and long-term health risks, outperforming traditional measures like HbA1c across diverse populations.
Key Takeaways
- •GluFormer, trained on over 10 million glucose measurements, provides generalizable representations for forecasting glycaemic parameters across various cohorts and conditions.
- •The model effectively stratifies individuals at risk for diabetes progression and cardiovascular mortality, with significant improvements over baseline HbA1c and CGM metrics.
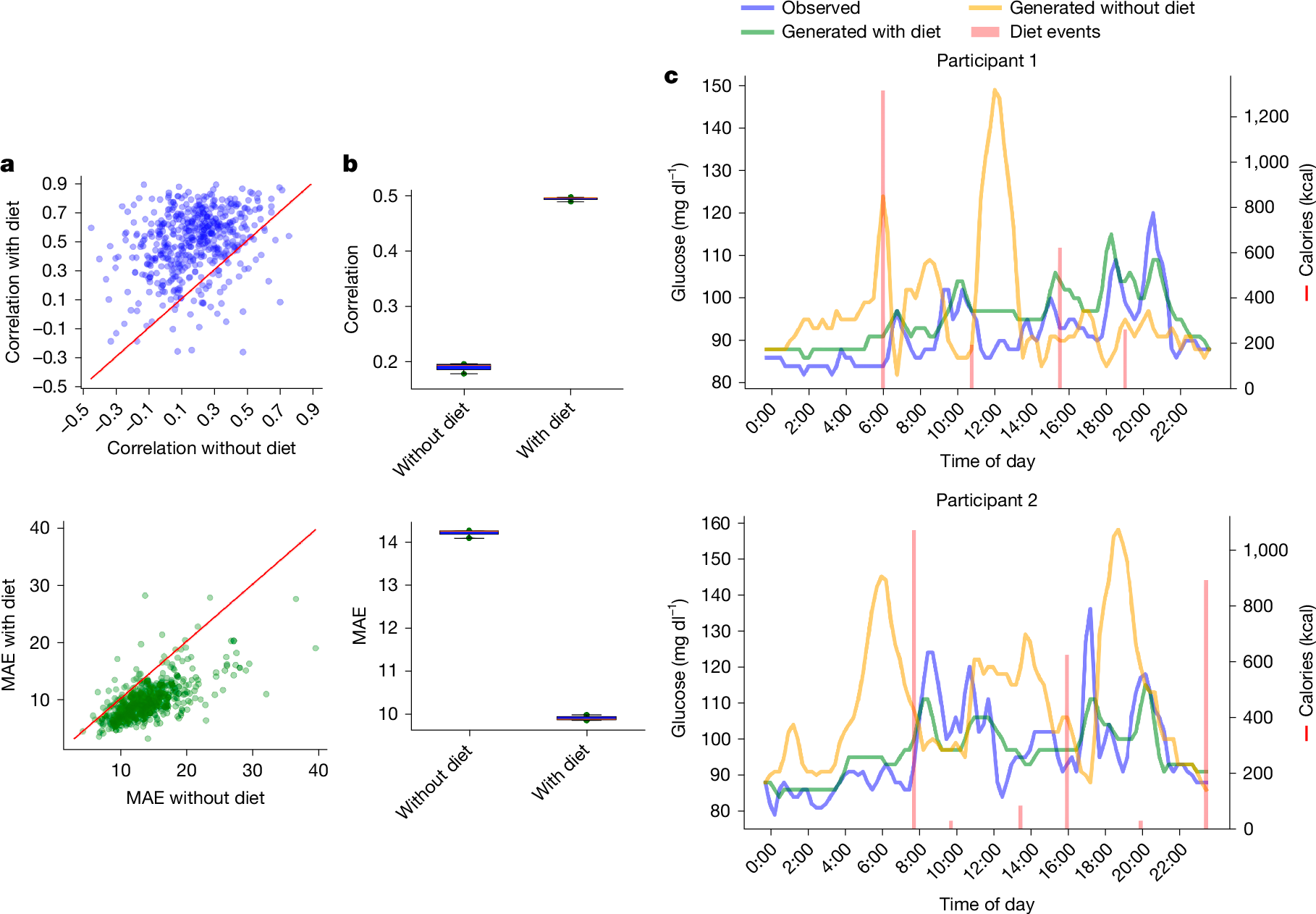
- •A multimodal extension integrating dietary data enhances prediction of individual glucose responses, supporting precision medicine approaches for metabolic health.
Tags
Abstract
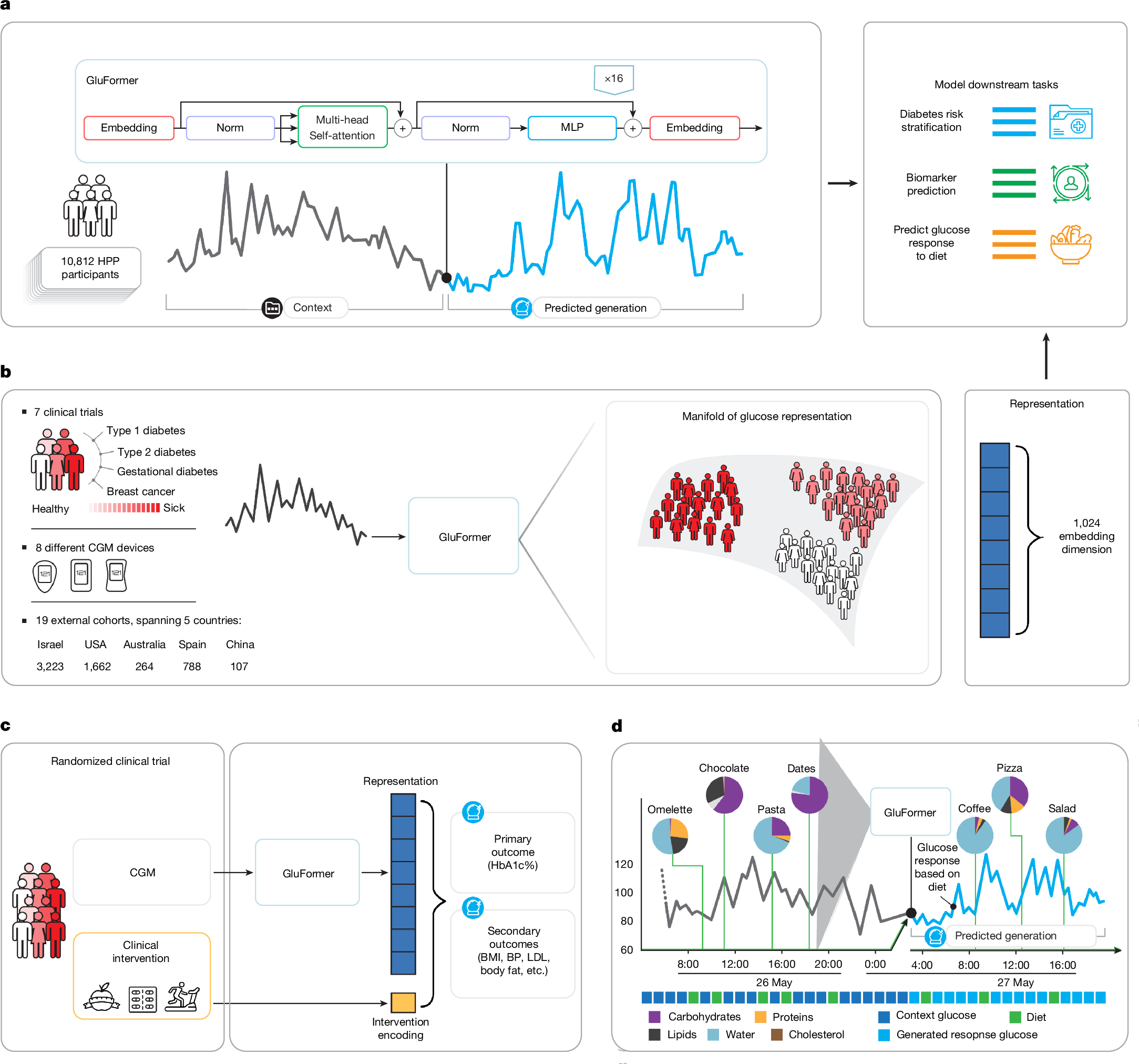
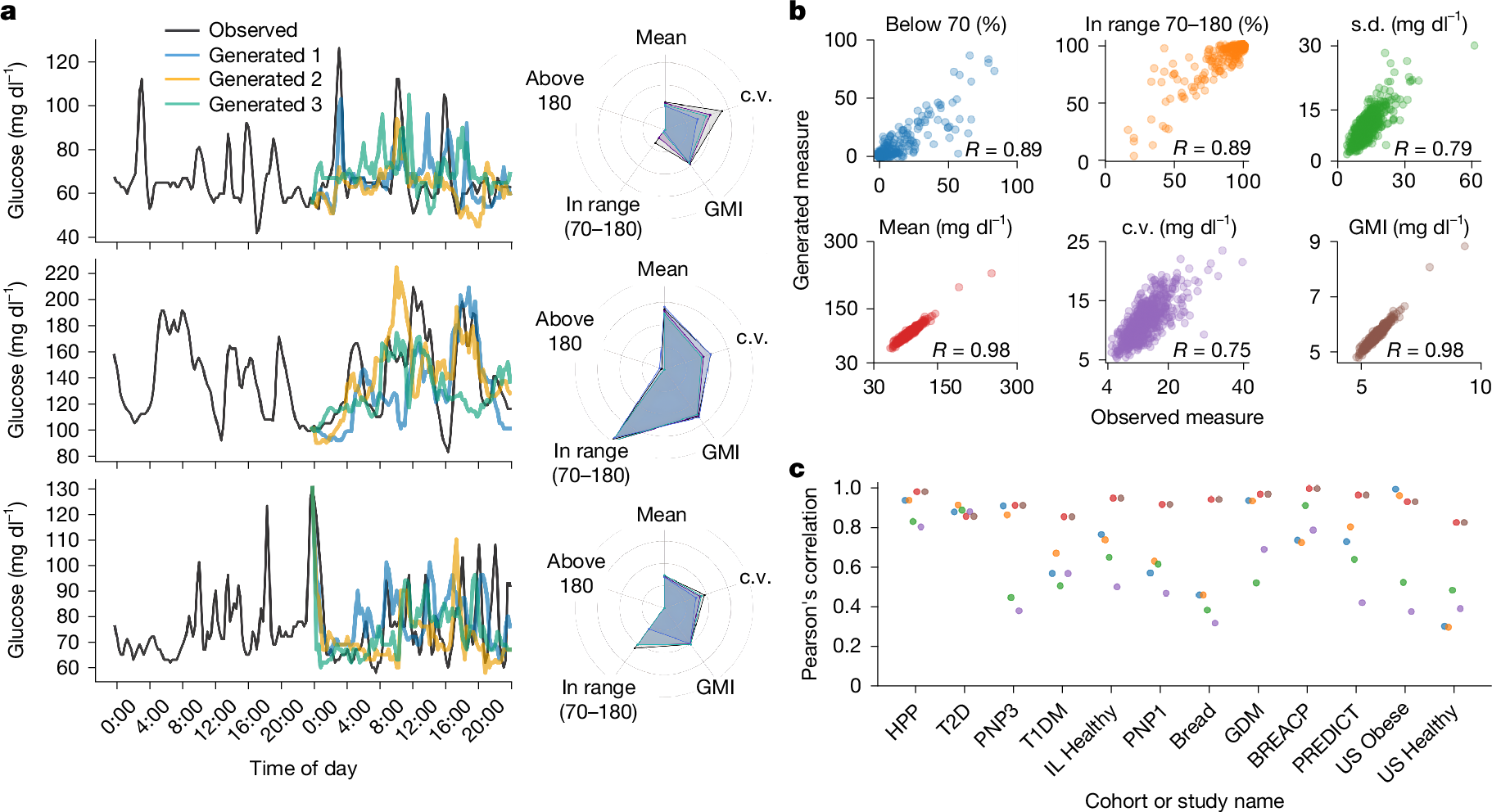
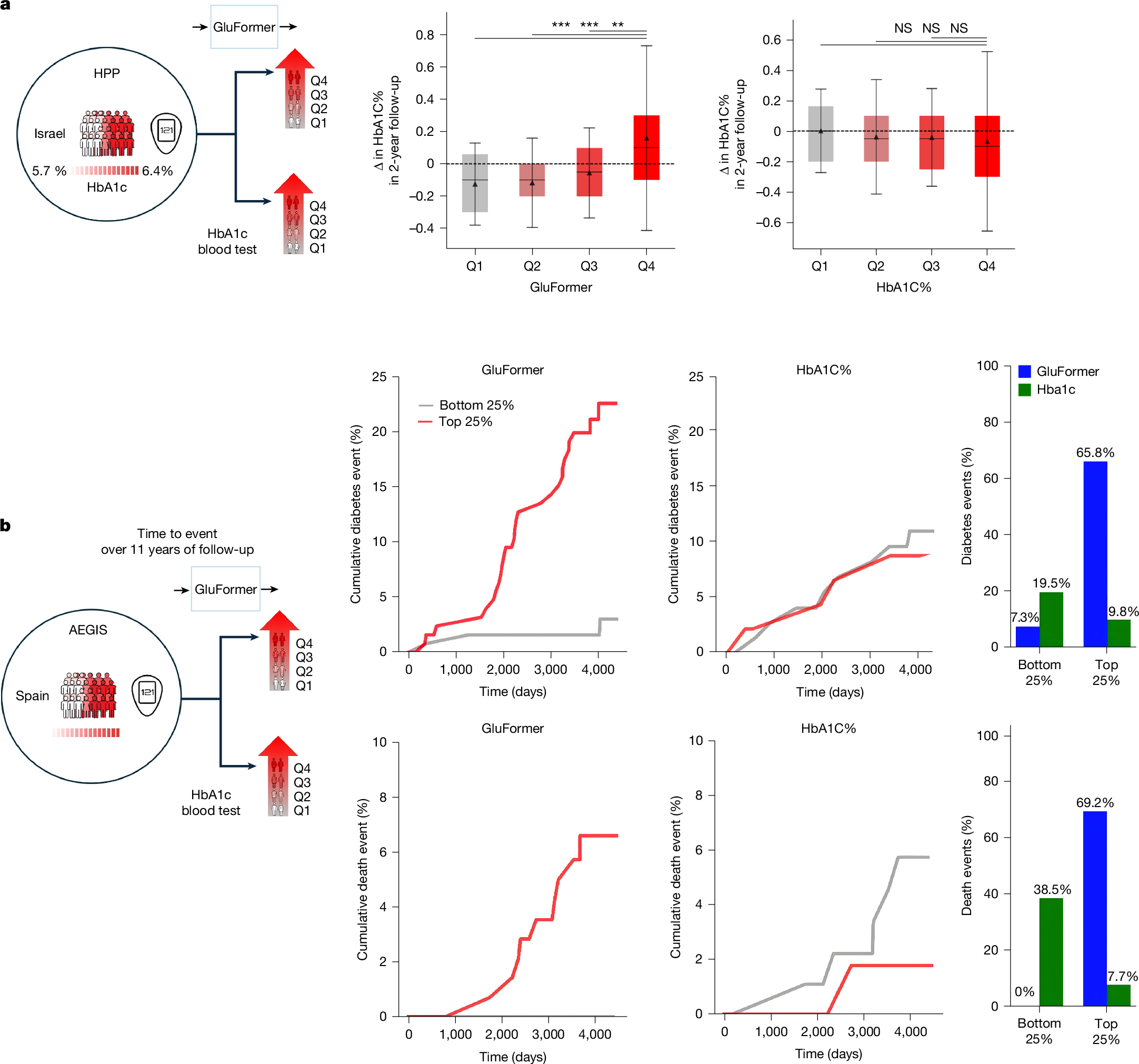
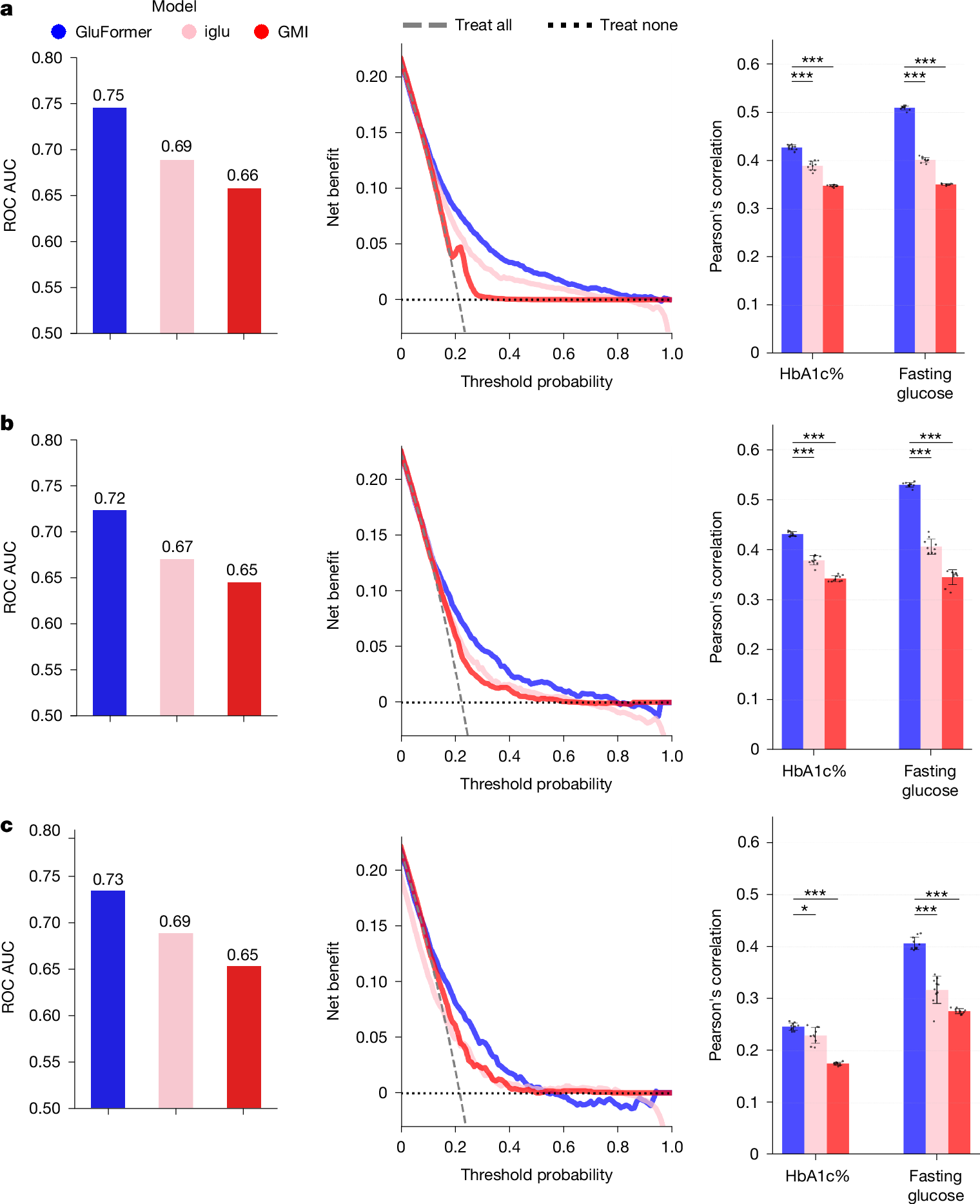
Continuous glucose monitoring (CGM) generates detailed temporal profiles of glucose dynamics, but its full potential for achieving glucose homeostasis and predicting long-term outcomes remains underutilized. Here we present GluFormer, a generative foundation model for CGM data trained with self-supervised learning on more than 10 million glucose measurements from 10,812 adults mainly without diabetes1,2. Using autoregressive prediction, the model learned representations that transferred across 19 external cohorts (n = 6,044) spanning 5 countries, 8 CGM devices and diverse pathophysiological states, including prediabetes, type 1 and type 2 diabetes, gestational diabetes and obesity. These representations provided consistent improvements over baseline blood glucose and HbA1c levels and other CGM-derived measures for forecasting glycaemic parameters3,4. In individuals with prediabetes, GluFormer stratified those likely to experience clinically significant increases in HbA1c over a 2-year period, outperforming baseline HbA1c and common CGM metrics. In a cohort of 580 adults with short-term CGM and a median follow-up of 11 years5, GluFormer identified individuals at elevated risk of diabetes and cardiovascular mortality more effectively than HbA1c. Specifically, 66% of incident diabetes cases and 69% of cardiovascular deaths occurred in the top risk quartile, compared with 7% and 0%, respectively, in the bottom quartile. In clinical trials, baseline CGM representations improved outcome prediction. A multimodal extension of the model that integrates dietary data generated plausible glucose trajectories and predicted individual glycaemic responses to food. Together, these findings indicate that GluFormer provides a generalizable framework for encoding glycaemic patterns and may inform precision medicine approaches for metabolic health.
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Data availability
The data used in this paper are part of the HPP and are accessible to researchers from universities and other research institutions (https://humanphenotypeproject.org/data-access). Interested bona fide researchers should contact [email protected] to obtain instructions for accessing the data. Deidentified participant data from the AEGIS study will be made available upon publication through the Runa Digital Repository (runa.sergas.gal). Access will require a signed data access agreement, and proposals should be directed to F.G.
Code availability
Implementation of GluFormer is available at GitHub (https://github.com/Guylu/GluFormer).
References
Shilo, S. et al. 10 K: a large-scale prospective longitudinal study in Israel. Eur. J. Epidemiol. 36, 1187–1194 (2021).
Reicher, L. et al. Deep phenotyping of health–disease continuum in the Human Phenotype Project. Nat. Med. 31, 3191–3203 (2025).
Nathan, D. M. et al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986 (1993).
King, P., Peacock, I. & Donnelly, R. The UK prospective diabetes study (UKPDS): clinical and therapeutic implications for type 2 diabetes. Br. J. Clin. Pharmacol. 48, 643–648 (1999).
Gude, F. et al. Glycemic variability and its association with demographics and lifestyles in a general adult population. J. Diabetes Sci. Technol. 11, 780–790 (2017).
Saab, K. et al. Capabilities of Gemini models in medicine. Preprint at https://doi.org/10.48550/arxiv.2404.18416 (2024).
Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature 622, 156–163 (2023).
Lutsker, G., Rossman, H., Godiva, N. & Segal, E. COMPRER: a multimodal multi-objective pretraining framework for enhanced medical image representation. Preprint at