N1-Methylpseudouridine directly modulates translation dynamics
TL;DR
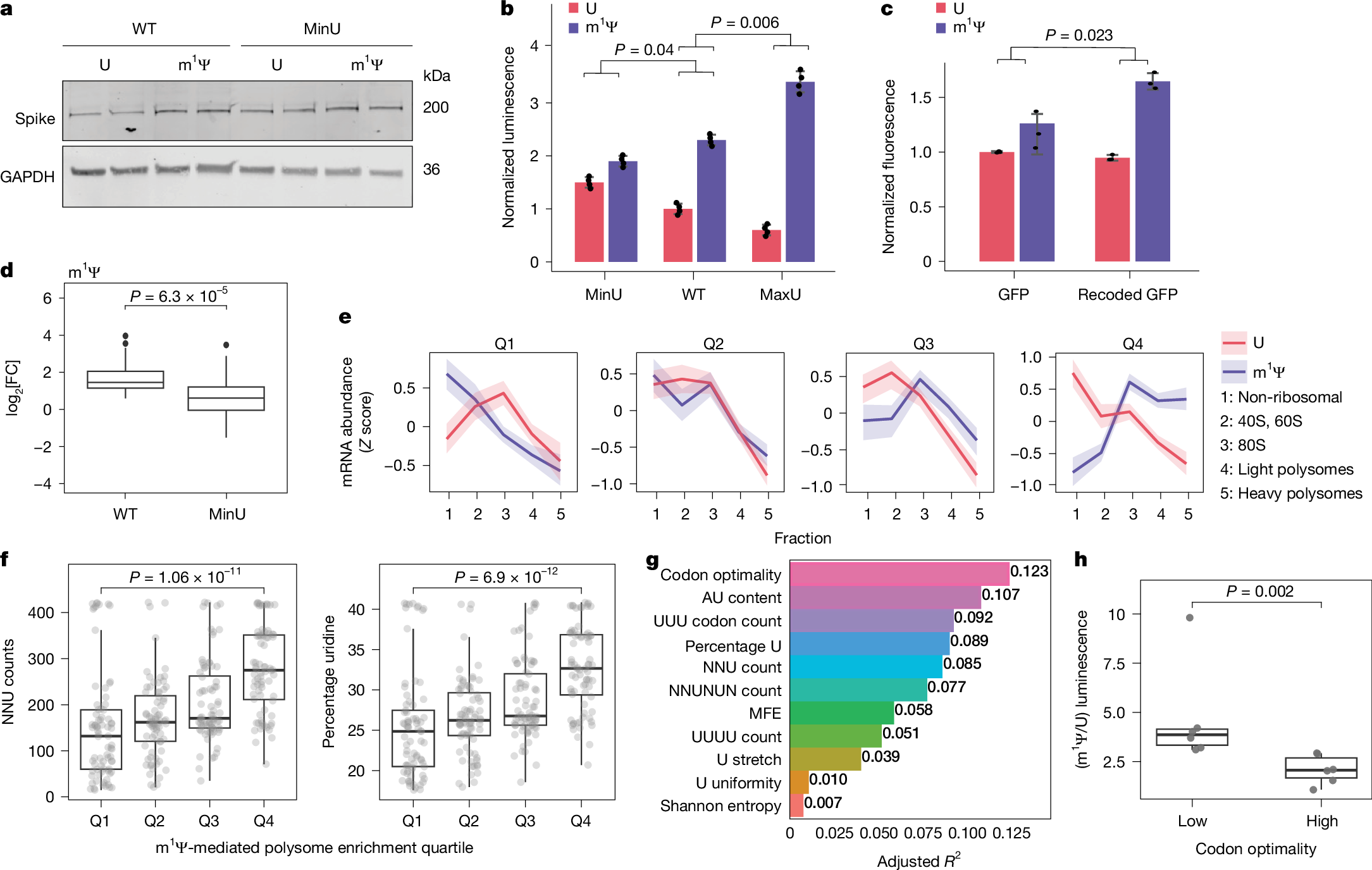
N1-methylpseudouridine (m1Ψ) in synthetic mRNA vaccines enhances protein production by slowing ribosome elongation in specific codon contexts while promoting translation initiation. Structural studies reveal m1Ψ alters ribosomal decoding center interactions, with strongest effects in mRNAs containing non-optimal codons with uridines at wobble positions.
Key Takeaways
- •m1Ψ increases ribosome density and protein production independent of immune activation mechanisms
- •m1Ψ directly slows ribosome movement in specific sequence contexts while enhancing translation initiation
- •Cryo-EM structural studies show m1Ψ alters interactions within the ribosomal decoding center
- •m1Ψ's enhancement effect is modulated by codon composition and strongest in mRNAs with non-optimal codons containing uridines at wobble positions
- •The findings demonstrate m1Ψ directly modulates translation dynamics to increase protein yield from synthetic mRNAs
Tags
Abstract
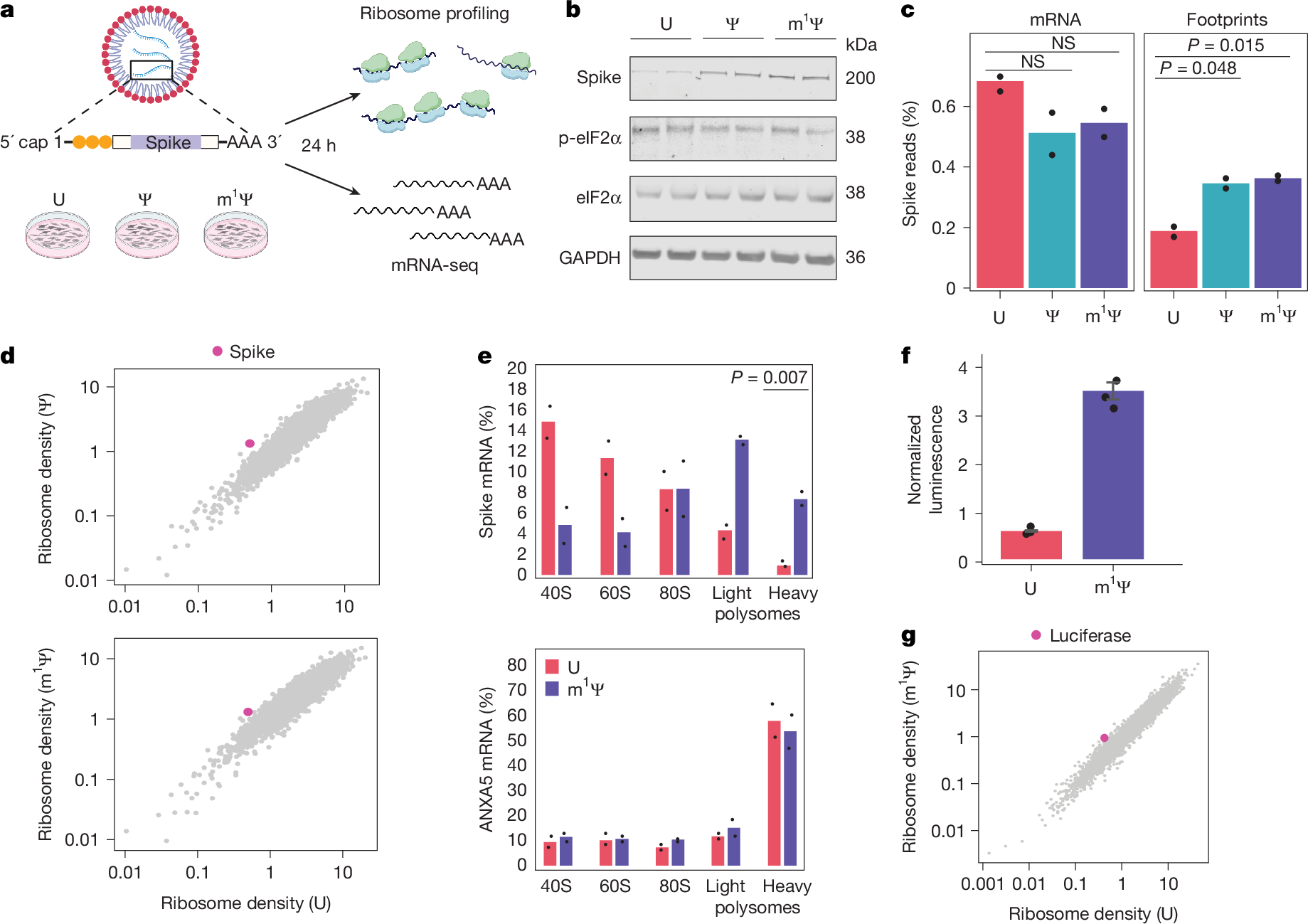
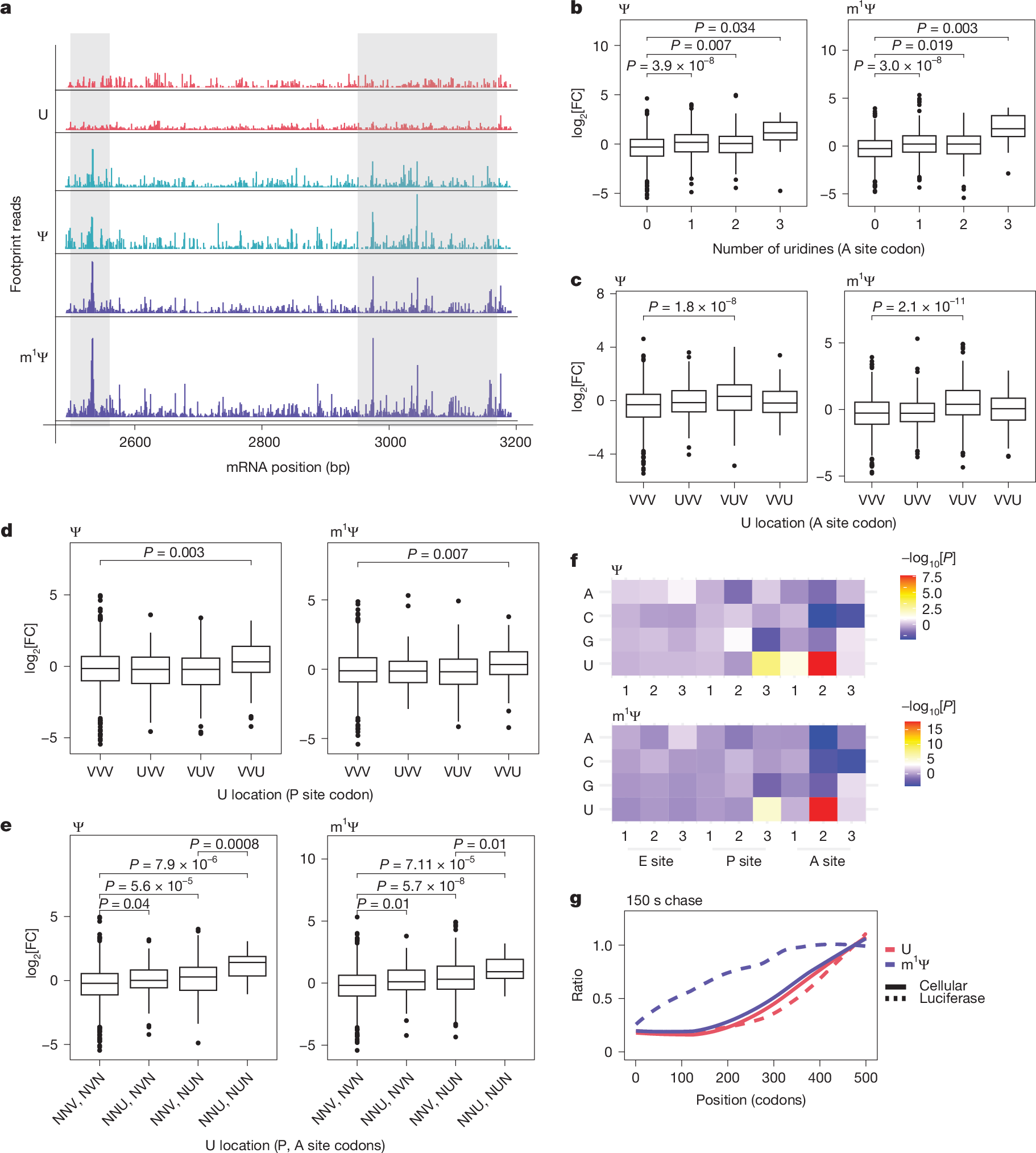
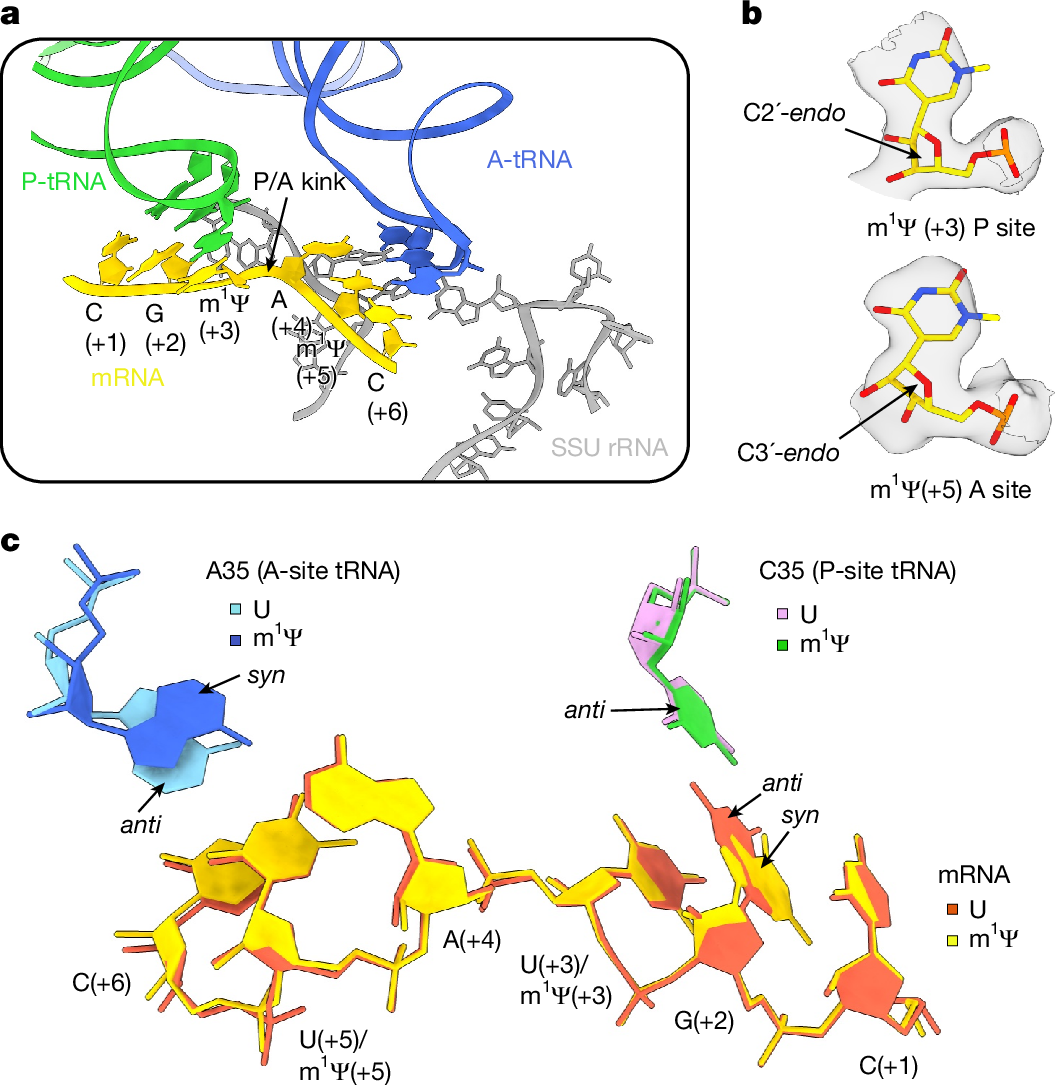
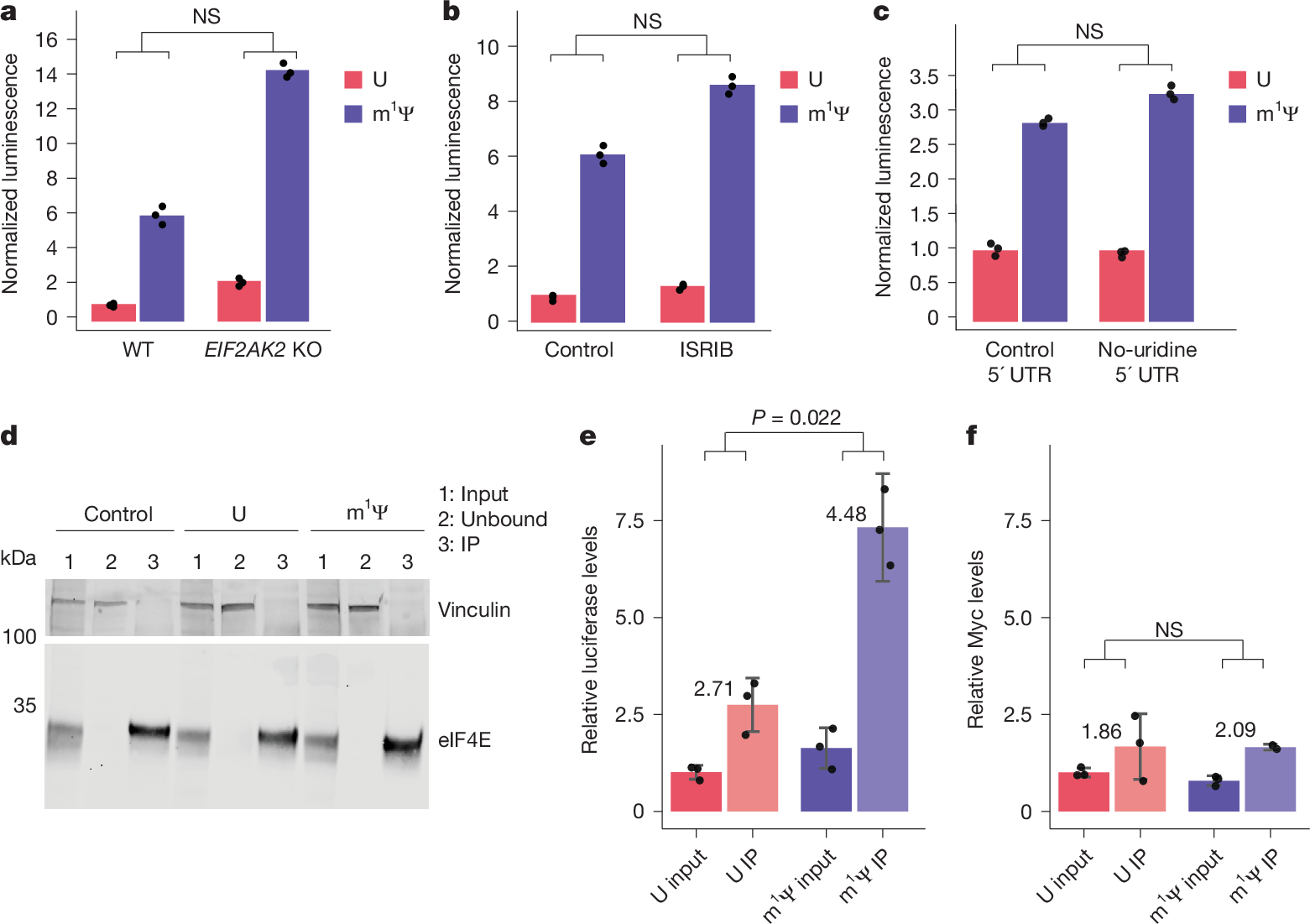
The considerable success of mRNA vaccines against SARS-CoV-2 has underscored the potential of synthetic mRNA as a transformative biomedical technology1. A critical feature of this approach is the incorporation of the modified nucleoside N1-methylpseudouridine (m1Ψ), which enhances antigen expression while reducing immunogenicity2,3,4,5. However, a comprehensive understanding of how m1Ψ influences translation remains incomplete. Here we use ribosome profiling at the subcodon resolution to show that m1Ψ increases ribosome density on synthetic mRNAs, leading to higher protein production independent of innate immune activation or eIF2α phosphorylation. We find that m1Ψ directly slows ribosome movement in defined sequence contexts while simultaneously promoting translation initiation. Structural studies using cryo-electron microscopy reveal that m1Ψ alters interactions within the ribosomal decoding centre, providing a mechanistic basis for slowed elongation. Furthermore, by introducing synonymous recoding that disrupts the modification-mediated changes in elongation, we show that the m1Ψ-dependent enhancement of protein output is modulated by codon composition, and that m1Ψ impact is strongest in mRNAs containing non-optimal codons with uridines at the wobble position. Together, these findings demonstrate that m1Ψ directly modulates translation dynamics, thereby increasing protein yield from synthetic mRNAs in specific sequence contexts.
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Data availability
All next-generation sequencing data files have been deposited at the Gene Expression Omnibus under accession GSE309271. The cryo-EM density maps of the ribosome complexes have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-55091 and EMD-55083. Atomic coordinates and structure factors have been deposited in the PDB under accession codes 9SPI and 9SPF.
References
Saxena, S. et al. The future of mRNA vaccines: potential beyond COVID-19. Cureus 17, e84529 (2025).
Anderson, B. R. et al. Nucleoside modifications in RNA limit activation of 2′−5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 39, 9329–9338 (2011).
Karikó, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Andries, O. et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015).
Anderson, B. R. et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010).
Bérouti, M. et al. Pseudouridine RNA avoids immune detection through impaired endolysosomal processing and TLR engagement. Cell 188, 4880–4895 (2025).
Cerneckis, J., Cui, Q., He, C., Yi, C. & Shi, Y. Decoding pseudouridine: an emerging target for therapeutic development. Trends Pharmacol. Sci. 43, 522–535 (2022).